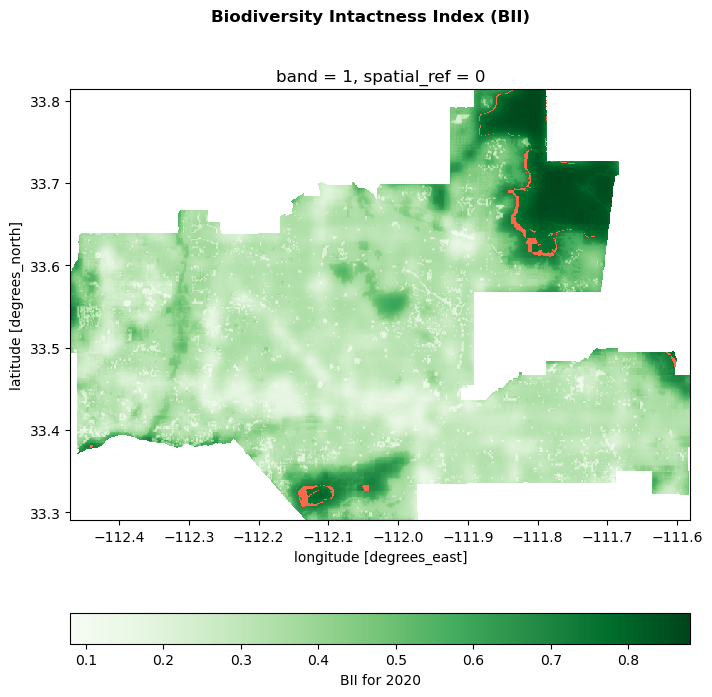
ID: io-biodiversity
Title: Biodiversity Intactness
Description: Generated by [Impact Observatory](https://www.impactobservatory.com/), in collaboration with [Vizzuality](https://www.vizzuality.com/), these datasets estimate terrestrial Biodiversity Intactness as 100-meter gridded maps for the years 2017-2020.
Maps depicting the intactness of global biodiversity have become a critical tool for spatial planning and management, monitoring the extent of biodiversity across Earth, and identifying critical remaining intact habitat. Yet, these maps are often years out of date by the time they are available to scientists and policy-makers. The datasets in this STAC Collection build on past studies that map Biodiversity Intactness using the [PREDICTS database](https://onlinelibrary.wiley.com/doi/full/10.1002/ece3.2579) of spatially referenced observations of biodiversity across 32,000 sites from over 750 studies. The approach differs from previous work by modeling the relationship between observed biodiversity metrics and contemporary, global, geospatial layers of human pressures, with the intention of providing a high resolution monitoring product into the future.
Biodiversity intactness is estimated as a combination of two metrics: Abundance, the quantity of individuals, and Compositional Similarity, how similar the composition of species is to an intact baseline. Linear mixed effects models are fit to estimate the predictive capacity of spatial datasets of human pressures on each of these metrics and project results spatially across the globe. These methods, as well as comparisons to other leading datasets and guidance on interpreting results, are further explained in a methods [white paper](https://ai4edatasetspublicassets.blob.core.windows.net/assets/pdfs/io-biodiversity/Biodiversity_Intactness_whitepaper.pdf) entitled “Global 100m Projections of Biodiversity Intactness for the years 2017-2020.”
All years are available under a Creative Commons BY-4.0 license.
Extent: <pystac.collection.Extent object at 0x0000016E9D485C10>
bii_2020_34.74464974521749_-115.38597824385106_cog {'datetime': None, 'proj:shape': [7992, 7992], 'end_datetime': '2020-12-31T23:59:59Z', 'proj:transform': [0.0008983152841195215, 0.0, -115.38597824385106, 0.0, -0.0008983152841195215, 34.74464974521749, 0.0, 0.0, 1.0], 'start_datetime': '2020-01-01T00:00:00Z', 'proj:code': 'EPSG:4326'}
bii_2019_34.74464974521749_-115.38597824385106_cog {'datetime': None, 'proj:shape': [7992, 7992], 'end_datetime': '2019-12-31T23:59:59Z', 'proj:transform': [0.0008983152841195215, 0.0, -115.38597824385106, 0.0, -0.0008983152841195215, 34.74464974521749, 0.0, 0.0, 1.0], 'start_datetime': '2019-01-01T00:00:00Z', 'proj:code': 'EPSG:4326'}
bii_2018_34.74464974521749_-115.38597824385106_cog {'datetime': None, 'proj:shape': [7992, 7992], 'end_datetime': '2018-12-31T23:59:59Z', 'proj:transform': [0.0008983152841195215, 0.0, -115.38597824385106, 0.0, -0.0008983152841195215, 34.74464974521749, 0.0, 0.0, 1.0], 'start_datetime': '2018-01-01T00:00:00Z', 'proj:code': 'EPSG:4326'}
bii_2017_34.74464974521749_-115.38597824385106_cog {'datetime': None, 'proj:shape': [7992, 7992], 'end_datetime': '2017-12-31T23:59:59Z', 'proj:transform': [0.0008983152841195215, 0.0, -115.38597824385106, 0.0, -0.0008983152841195215, 34.74464974521749, 0.0, 0.0, 1.0], 'start_datetime': '2017-01-01T00:00:00Z', 'proj:code': 'EPSG:4326'}